Asim Iqbal, PhD

Machine Learning | Neuroscience
Email: iqbala@ini.ethz.ch
About Me:
With a background in Machine Learning and Neuroscience, I aim to understand the anatomical and functional mysteries of brain to build robust solutions for brain disorders.
Affiliation:
I serve as a Visiting Research Scientist at the Brain Research Institute (HiFo) and Neuroscience Center Zurich (ZNZ), ![]() where I assist in uncovering computational principles in the developing mouse cortex through deep generative models.
where I assist in uncovering computational principles in the developing mouse cortex through deep generative models.
As an Affiliate Faculty, I co-advise a small research group in my free time at the Department of Computer Science, ![]() and
and ![]() . We develop high-performing deep neural networks inspired by the mouse visual cortex and domain adaptive deep learning models for neuroimaging datasets.
. We develop high-performing deep neural networks inspired by the mouse visual cortex and domain adaptive deep learning models for neuroimaging datasets.
I also serve as a Scientific Advisory Board Member in a few tech startups focusing on Computer Vision and Machine Learning such as Wizlabs and LAAM.
Education:
I worked as a Postdoctoral Research Scientist (2020-2021) on the intersection of AI and Neuroscience in Mathis Lab, Center for Intelligent Systems (CIS) | ELLIS Unit, Swiss Federal Institute of Technology ![]() .
.
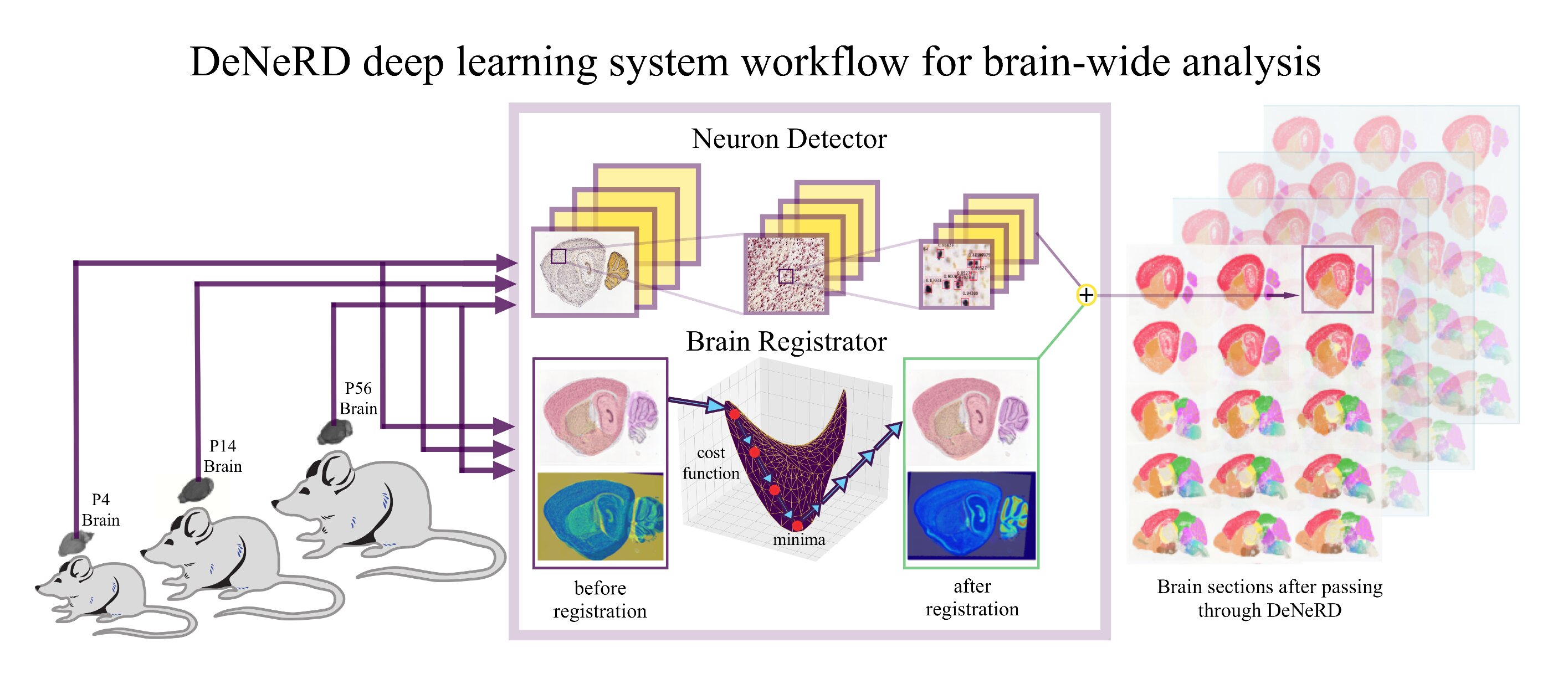
Prior to this, I completed my PhD (Fall 2016 - Fall 2019) in the field of Computational Neuroscience and Machine Learning on the topic: 'Exploring the principles of brain-wide development through deep learning' at Institut für Hirnforschung (HiFo) and Zentrum für Neurowissenschaften Zürich (ZNZ), ![]() . My PhD thesis was advised by Prof. Theofanis Karayannis along with Prof. Fritjof Helmchen and Prof. Mehmet Fatih Yanik as advisory committe members. The focus of my PhD project was to develop deep learning-based tools to analyse neuro-imaging datasets which includes detection of neurons and generating an automated atlas for developing mouse and human brain sections captured through various imaging modalities. Morover, I worked on explaining the functional and anatomical correlates of neural responses in the mouse somatosensory and visual cortex.
. My PhD thesis was advised by Prof. Theofanis Karayannis along with Prof. Fritjof Helmchen and Prof. Mehmet Fatih Yanik as advisory committe members. The focus of my PhD project was to develop deep learning-based tools to analyse neuro-imaging datasets which includes detection of neurons and generating an automated atlas for developing mouse and human brain sections captured through various imaging modalities. Morover, I worked on explaining the functional and anatomical correlates of neural responses in the mouse somatosensory and visual cortex.
Before starting my PhD, I studied Electrical Engineering (2006-2010) as an undergrad followed by a Masters (2013-2015) in Neural Systems and Computation at the Institute of Neuroinformatics (
![]() ), Department of IT and Electrical Engineering (D-ITET), ETH Zürich and worked on 'Enhancing scale-invariance in a convolutional neural network' as a thesis project by introducing a neuro-inspired layer in the deep neural network that mimics the functionality of complex cells in the visual cortex. Moreover, I developed a robust framework to detect very fast moving objects in real-time through frame-based running deep neural network with an interface to a spike-based running neuromorphic retina sensor.
), Department of IT and Electrical Engineering (D-ITET), ETH Zürich and worked on 'Enhancing scale-invariance in a convolutional neural network' as a thesis project by introducing a neuro-inspired layer in the deep neural network that mimics the functionality of complex cells in the visual cortex. Moreover, I developed a robust framework to detect very fast moving objects in real-time through frame-based running deep neural network with an interface to a spike-based running neuromorphic retina sensor.
Research Experience:
In past, I worked as a Lead Machine Learning Scientist (2021-2022) at ![]() for discovering novel solutions for neurodegenerative disorders through AI-based drug discovery.
for discovering novel solutions for neurodegenerative disorders through AI-based drug discovery.
From Summer 2019 to Spring 2020, I worked as a Machine Learning Scientist Intern on a NeuroMoonshot (Project Amber) at
![]()
![]() - The Moonshot Factory, Mountain View, California.
- The Moonshot Factory, Mountain View, California.
In Summer of 2018, I attended Summer Workshop on the Dynamic Brain (SWDB) at
and
![]() in Seattle, Washington and worked on developing a deep neural network-based decoder to predict neural responses in mouse visual cortex.
in Seattle, Washington and worked on developing a deep neural network-based decoder to predict neural responses in mouse visual cortex.
In Spring of 2017, I attended Connectomics at Neuroscience School of Advancted Studies
![]() in Siena, Italy.
in Siena, Italy.
In Summer of 2016, I worked as a Research Intern with Brain-inspired Computing Team on classification of hand gestures through deep learning and its implementation on TrueNorth neuromorphic chip at
![]() - Almaden, San Jose, California.
- Almaden, San Jose, California.
From Spring 2015 to Spring 2016, I worked as a Visiting Researcher with Prof. Jim DiCarlo in Brain and Cognitive Sciences Department,
![]() at Cambridge, Massachusetts. I worked on predicting neural responses in primate visual cortex through performance optimized deep neural networks.
at Cambridge, Massachusetts. I worked on predicting neural responses in primate visual cortex through performance optimized deep neural networks.
From 2013 to 2014, I worked as a Research Assistant in Prof. Fritjof Helmchen's lab at the Brain Research Institute,
![]() . I worked on developing algorithms to analyse whisker detection in mouse somatosensory tasks through LeapMotion device.
. I worked on developing algorithms to analyse whisker detection in mouse somatosensory tasks through LeapMotion device.
Research Interests:
My research interests are broadly categorized into the following:
- Neuro-inspired Artificial Intelligence
- Domain Generalization
Publications:
Payette, Kelly, Priscille de Dumast, Hamza Kebiri, Ivan Ezhov, Johannes C. Paetzold, Suprosanna Shit, Asim Iqbal et al. "An automatic multi-tissue human fetal brain segmentation benchmark using the Fetal Tissue Annotation Dataset." Nature Scientific Data 8, 167 (2021).[pdf]
Rahel Kastli*, Rasmus Vighagen*, Alexander van der Bourg*, Ali Ozgur Argunsah*, Asim Iqbal, Fabian F. Voigt, Daniel Kirschenbaum, Adriano Aguzzi, Fritjof Helmchen, and Theofanis Karayannis. "Developmental divergence of sensory stimulus representation in cortical interneurons." Nature Communications 11, 5729 (2020).[pdf]
Hassan, Mahmood, Asim Iqbal, and S. M. Shamsul Islam. "Exploring Intensity Invariance in Deep Neural Networks for Brain Image Registration." Digital Image Computing: Techniques and Applications (DICTA), Melbourne, Australia, 2020.[pdf]
Asim Iqbal, Asfandyar Sheikh, and Theofanis Karayannis. "DeNeRD: high-throughput detection of neurons for brain-wide analysis with deep learning." Scientific Reports 9, no. 1 (2019): 1-13.[pdf]
Asim Iqbal, Phil Dong, Christopher M. Kim, and Heeun Jang. "Decoding neural responses in mouse visual cortex through a deep neural network." In International Joint Conference on Neural Networks (IJCNN), pp. 1-7. IEEE, 2019. [pdf]
Asim Iqbal (featured) "A deeply learned brain atlas." Nature Methods (2019): 680-680.[pdf]
Asim Iqbal, Romesa Khan, and Theofanis Karayannis. "Developing a brain atlas through deep learning." Nature Machine Intelligence 1, no. 6 (2019): 277-287.[pdf]
Asim Iqbal, Asfandyar Sheikh, and Theofanis Karayannis. "Exploring brain-wide development of inhibition through deep learning. arXiv (2018).[pdf]
Research Projects:
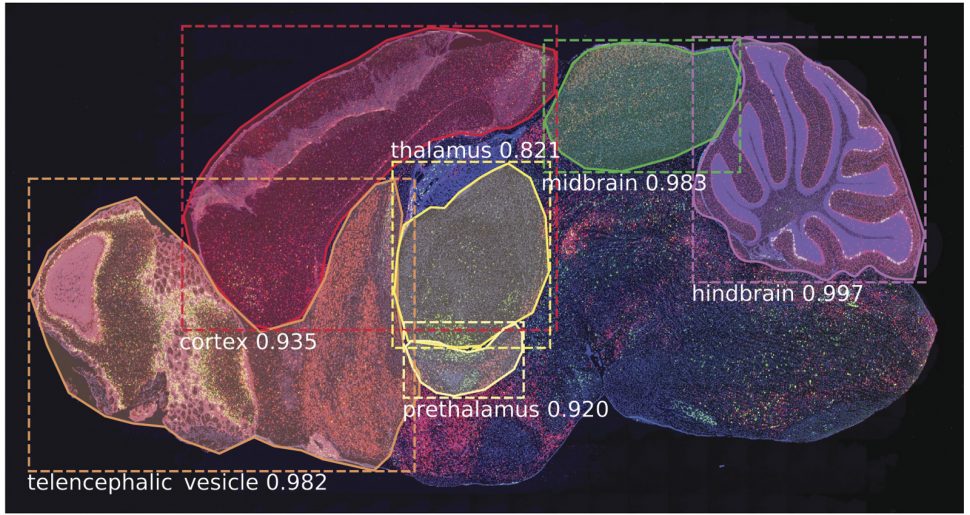
[SeBRe]: Segmenting Brain Regions with deep learning
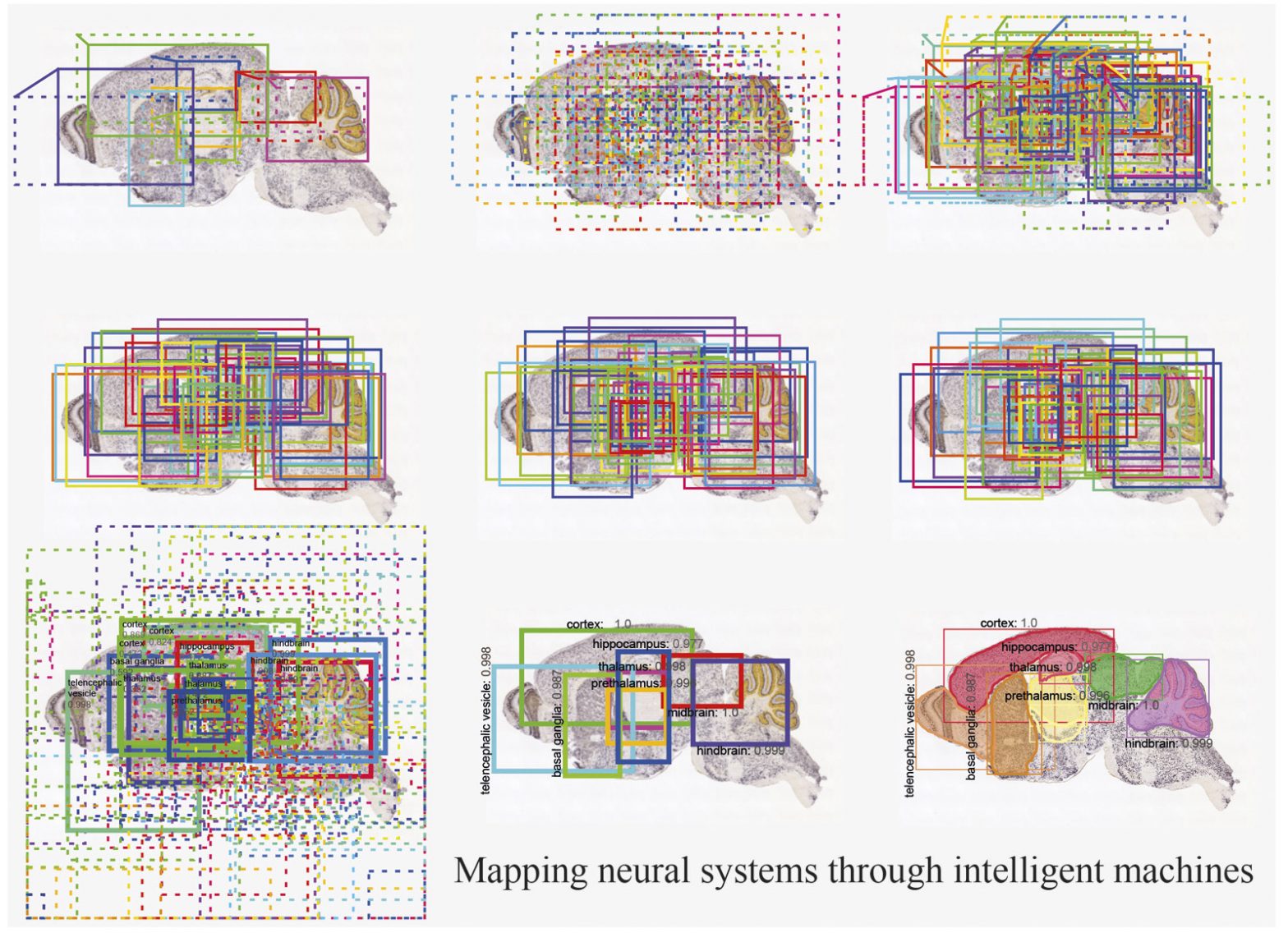
One of the main challenges faced by biologists in general and neuroscientists in partcular is to register mouse/human brain section images to a standard reference atlas. It requires an anatomical expert with a basic training to recognise and precisely annotate brain regions which is a very time consuming and exhausting process. Although, transformation algorithms have tackeled the automated brain image registration problem to a large extent but they still require fine tuning of parameters for any unseen brain section image. This fine tuning process easily takes hours as these algorithms are just trying to minimise a cost function to match the input image to a reference image. Furthermore, these algorithms are prone to errors and cannot simply work if any brain region is missing or distorted. We introduce a concept of registration through segmentation: we train a deep learning model for instance segmentation to classify and segment all the brain regions in a given image with high accuracy. This approach is independent of data modality and can also serve to help generate an atlas for animal brain ages on which no anatomical atlas is pre-existed. Hence, developing a brain atlas through deep learning. Our study is published in Nature Machine Intelligence and made it to the cover of the journal. [paper] [code]
Asim Iqbal, Romesa Khan, and Theofanis Karayannis. "Developing a brain atlas through deep learning."
Nature Machine Intelligence 1.6 (2019): 277-287.
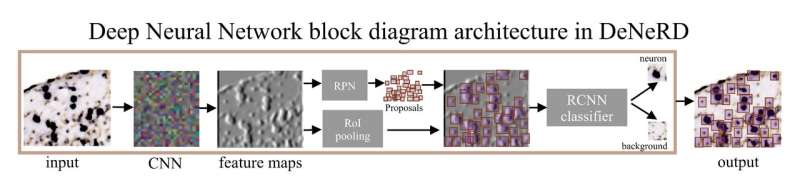
[DeNeRD]: Detecting Neurons in Regions of Development with deep learning
Neurons are captured with variability in shape, size, structure, intensity, etc. which becomes a challenge to detect them for high-throughput analysis of brain section images. We consider it as an object classification and detection problem and generated a ground-truth dataset of thousands of labelled neurons from mouse brain with different imaging modalities and genetic markers. We develop a deep neural network-based architecture to detect neurons in the entire 2D brain sections with high precision. Our study is published in Scientific Reports and made it to the Top 100 in Neuroscience. [paper] [code]
Asim Iqbal, Asfandyar Sheikh, and Theofanis Karayannis. "DeNeRD: high-throughput detection of neurons for brain-wide analysis with deep learning."
Scientific Reports 9.1 (2019): 1-13.
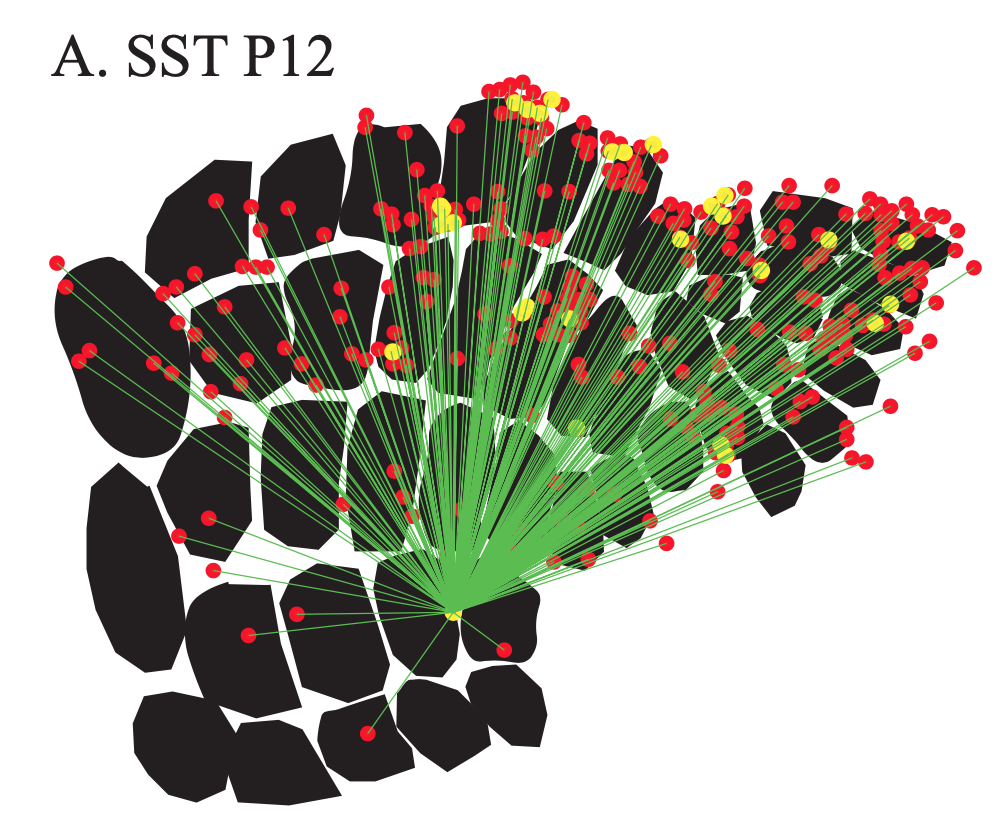
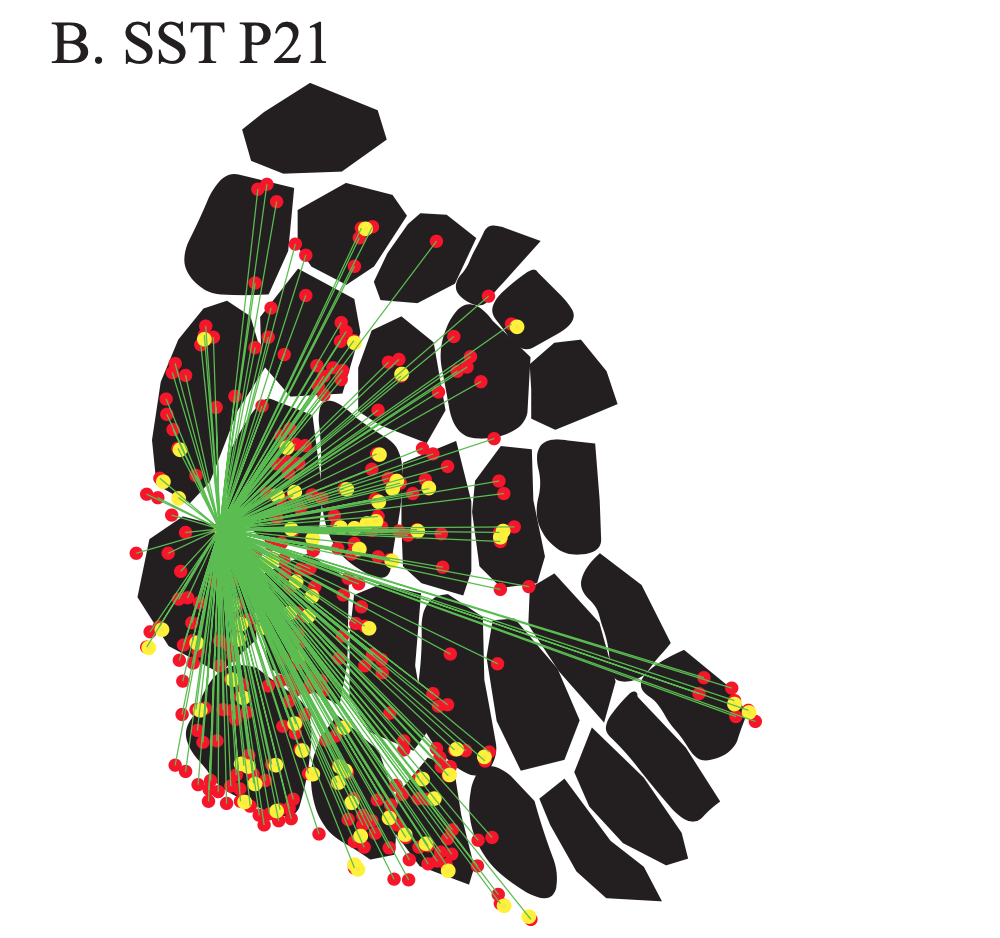
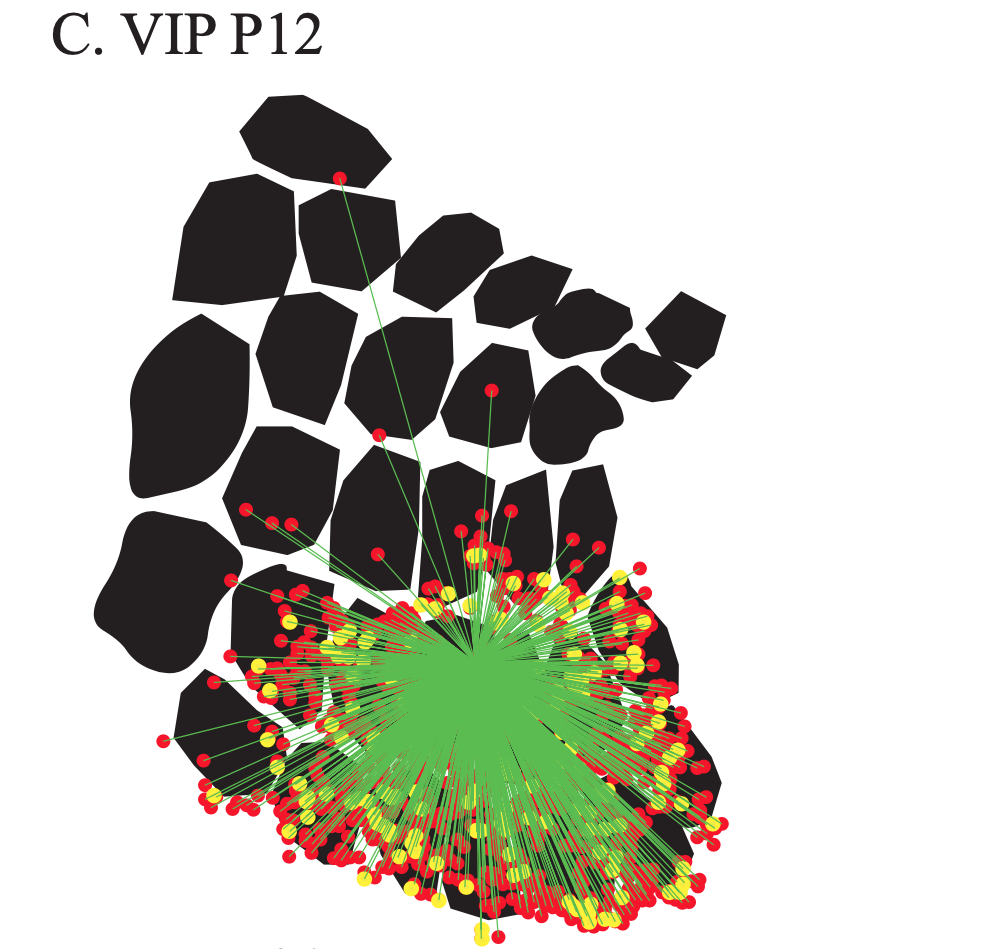
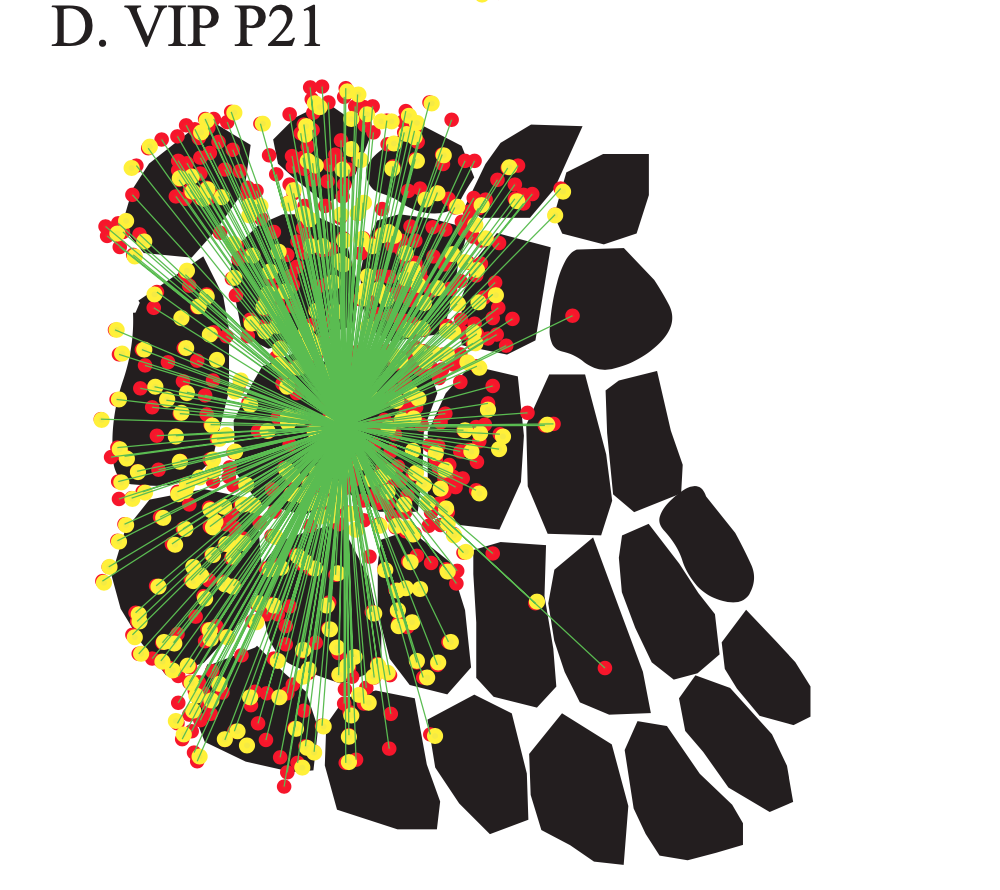
Exploring the functional and anatomical motifs of developing mouse somatosensory cortex
Two inhibitory cell types involved in modulating barrel cortex activity and perception during active whisking in adult mice, are the VIP+ and SST+ interneurons. Here we identify a developmental transition point of structural and functional rearrangements onto these interneuron types around the start of active sensation at P14. Using in vivo two-photon Ca2+ imaging, we find that before P14, both interneuron types respond stronger to a multi-whisker stimulus, whereas after P14 their responses diverge, with VIP+ cells losing their multi-whisker preference and SST+ neurons enhancing theirs. Rabies virus tracings followed by tissue clearing, as well as photostimulation-coupled electrophysiology reveal that SST+ cells receive higher cross-barrel inputs compared to VIP+ at both time points. In addition, we also uncover that whereas prior to P14 both cell types receive direct input from the sensory thalamus, after P14 VIP+ cells show reduced inputs and SST+ cells largely shift to motor-related thalamic nuclei. Our study is published in Nature Communications. [paper]
Rahel Kastli, Rasmus Vighagen, Alexander van der Bourg, Ali Ozgur Argunsah,
Asim Iqbal, Fabian F. Voigt, Daniel Kirschenbaum, Adriano Aguzzi, Fritjof Helmchen, and Theofanis Karayannis.
"Developmental Divergence of Sensory Stimulus Representation in Cortical Interneurons." Nature Communications (2020).
Code:
1. Developing a brain atlas through deep learning
2. DeNeRD: high-throughput detection of neurons for brain-wide analysis with deep learning
News:
-
I participated in the 'Panel Discussion with Interns at Google' as an invited speaker at the event of Google Developers Student Clubs, organised by ETH Zurich and Kings College London.

-
I gave a talk on 'Mapping Neural Development through Artificial Intelligence' as an invited speaker at the SBASSE, LUMS

-
My submission on 'Deep Clear Neural Art' won the Annual Grand Prize in 2021 NeuroArt Image Contest 🕸🎨🥇
-
Starting Fall 2021, I am joining Cajal Neuroscience as a Lead Machine Learning Scientist to discover novel solutions for neurodegenerative disorders through Computational Neuroimaging and AI.
-
I joined the SCI-AI Podcast to discuss 'How do AI and Neuroscience benefit each other?'

-
I am invited as a speaker at the ZNZ Symposium 2021
 in the Machine learning in Neuroscience and Neuroimaging workshop, organised by the University of Zurich and ETH Zurich.
in the Machine learning in Neuroscience and Neuroimaging workshop, organised by the University of Zurich and ETH Zurich. -
Our paper An automatic multi-tissue human fetal brain segmentation benchmark using the Fetal Tissue Annotation Dataset
 is featured in the Fetal Tissue Annotation Challenge by MICCAI 2021, sponsored by NVIDIA.
is featured in the Fetal Tissue Annotation Challenge by MICCAI 2021, sponsored by NVIDIA. -
Our paper Developmental divergence of sensory stimulus representation in cortical interneurons
 is featured by Nature Communications as a spotlight in From Brain to Behaviour.
is featured by Nature Communications as a spotlight in From Brain to Behaviour. -
Starting Summer 2020, I am joining Mathis Lab
 at Swiss Federal Institute of Technology (EPFL) as a Postdoctoral Fellow to work on Neuro-inspired AI systems.
at Swiss Federal Institute of Technology (EPFL) as a Postdoctoral Fellow to work on Neuro-inspired AI systems. -
LUMS
 featured my talk on Exploring the computational principles of brain-wide development through deep learning in SBASSE Newsletter 2020.
featured my talk on Exploring the computational principles of brain-wide development through deep learning in SBASSE Newsletter 2020. -
DeNeRD
 is featured by Scientific Reports in Top 100 in Neuroscience.
is featured by Scientific Reports in Top 100 in Neuroscience. -
DeNeRD
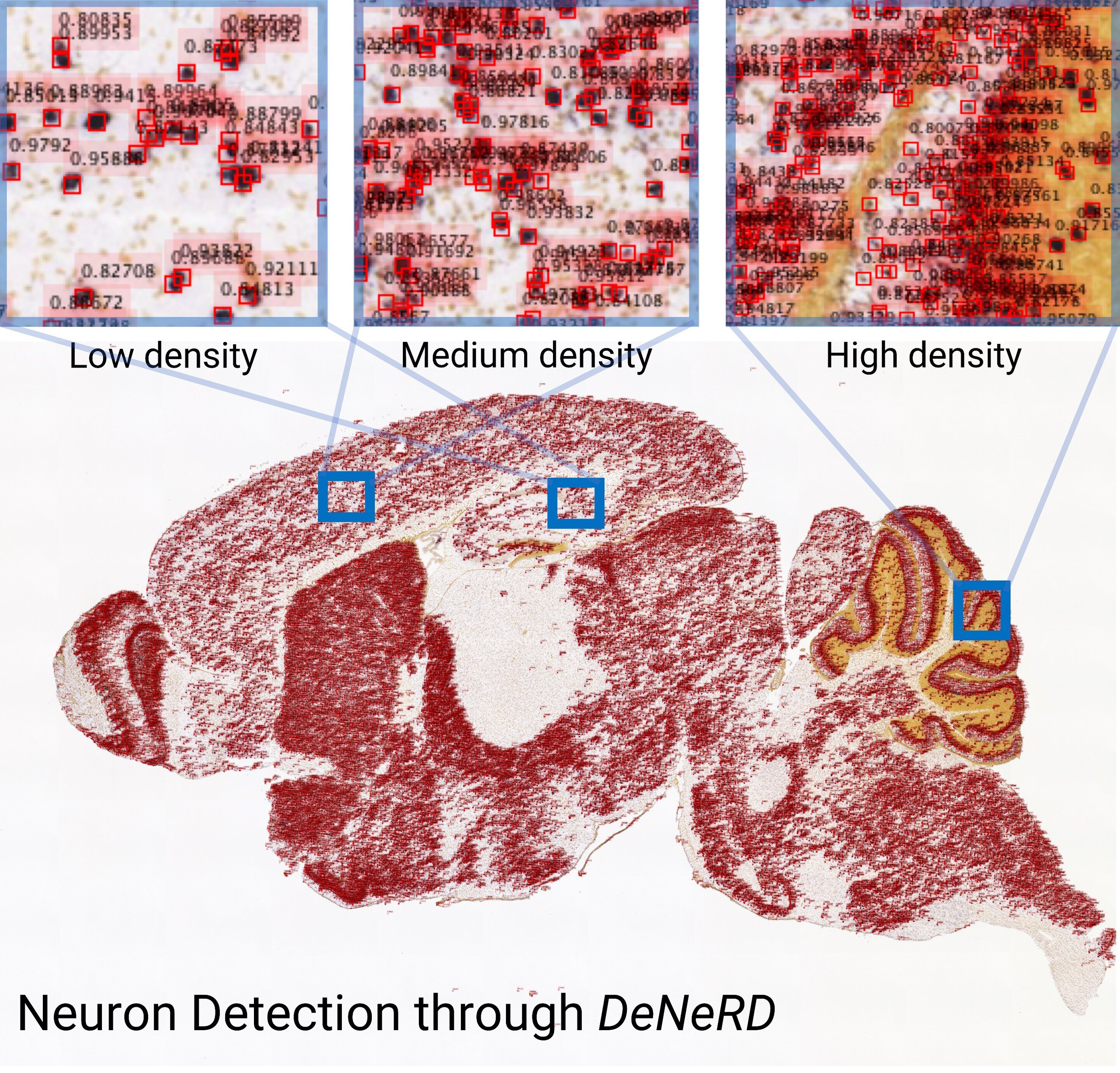 is featured by Medical Xpress in DeNeRD: an AI-based method to process whole images of the brain.
is featured by Medical Xpress in DeNeRD: an AI-based method to process whole images of the brain. -
SeBRe
 is featured by Singularity Hub in Three Invaluable Ways AI and Neuroscience Are Driving Each Other Forward.
is featured by Singularity Hub in Three Invaluable Ways AI and Neuroscience Are Driving Each Other Forward. -
SeBRe
 is featured by Nature Methods in A deeply learned brain atlas.
is featured by Nature Methods in A deeply learned brain atlas. -
SeBRe
 is featured by SciGlow in Developing brain maps through artificial intelligence.
is featured by SciGlow in Developing brain maps through artificial intelligence. -
SeBRe
 is featured by Swiss Cognitive in Thanks to deep learning, the tricky business of making brain atlases just got a lot easier.
is featured by Swiss Cognitive in Thanks to deep learning, the tricky business of making brain atlases just got a lot easier. -
SeBRe
 is featured by Singularity Hub in How Deep Learning Is Transforming Brain Mapping.
is featured by Singularity Hub in How Deep Learning Is Transforming Brain Mapping. -
SeBRe
 is featured by Tech Xplore in Developing brain atlas using deep learning algorithms.
is featured by Tech Xplore in Developing brain atlas using deep learning algorithms.
Teaching:
Courses Taught:
- Neuroscience Block Course (Spring 2019), UZH/ETH Zurich
- Programming in Biology (Fall 2018), UZH/ETH Zurich
- Neuroscience Block Course (Spring 2017), UZH/ETH Zurich
Current Students:
- Eunseo Sung (M.Sc. Comp Bio, Cornell University) - AI Intern
- Abdul Basit (M.Sc. Computer Science, LUMS) - Master Thesis
Supervised (Past) Students:
- Taha Razzaq (M.Sc. Computer Science, LUMS) - Master Thesis
- Hisan Naeem (B.Sc. Computer Science, LUMS) - Bachelor Thesis
- Ahmed Qazi (B.Sc. Computer Science, LUMS) - Bachelor Thesis
- Hamd Jalil (B.Sc. Computer Science, LUMS) - Bachelor Thesis
- Valentin Bruttin (M.Sc. Life Sciences & Engineering, EPFL) - Master Thesis
- Ella McPherson (M.Sc. Health Sciences and Technology, ETH Zurich) - Comp Neuro Intern
- Ali Bukhari (B.Sc. Computer Science, LUMS) - Bachelor Thesis
- Hamza Khalid (B.Sc. Computer Science, LUMS) - Internship
- Namra Aamir (B.Sc. Biology, LUMS) - Comp Neuro Intern
- Romesa Khan (M.Sc. Neuroscience, ETH Zurich) - Master Thesis
- Asfandyar Shiekh (B.Sc. Electrical Engineering, ETH Zurich) - ML Intern
- Markus Suter (M.Sc. Neuroscience, UZH) Master Thesis Co-supervised
- Laurens Bohlen (M.Sc. Neuroscience, UZH) Neuroscience Block Course
- Victor Ibañez (M.Sc. Neuroscience, UZH) Neuroscience Block Course
- Andrin Abegg (M.Sc. Neuroscience, ETH Zurich) Neuroscience Block Course
- Michel Schmidlin (M.Sc. Neuroscience, ETH Zurich) Neuroscience Block Course
Interested Students:
Any undergrad or graduate student in Computer Science (or similar STEM field) at UZH/ETH Zurich/LUMS interested in working with me on topics related to Neuro-inspired Artificial Intelligence then drop me an email:
iqbala{at}ini.ethz.ch
Thoughts:
-
How to survive in Pakistan amidst Coronavirus pandemic?
 My thoughts on Medium about the alarming Covid-19 situation in Pakistan.
My thoughts on Medium about the alarming Covid-19 situation in Pakistan. -
My thoughts on 'How do AI and Neuroscience benefit each other?' on SCI-AI Podcast.